Author Contributions
Academic Editor: Hem D Shukla, Professor in Department of Biology at University of Maryland, USA.
Checked for plagiarism: Yes
Review by: Single-blind
Copyright © 2018 Leonid Tarassishin
 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests
The authors have declared that no competing interests exist.
Citation:
Introduction
Biomarkers represent a significant area of research and clinical applications. Pub-Med search have shown more than 879,000 publications that used word “Biomarker”. The global biomarkers market is estimated to reach USD 53.34 Billion by 2021 from USD 27.95 Billion in 2016, at a CAGR (compound annual growth rate) of 13.8% from 2016 to 2021*.
* By: marketsandmarkets.com; Publishing Date: February 2017; Report Code: BT 2120
What are biomarkers? As stated by the definition of National Institute of Health “Biomarker is a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacological responses to a therapeutic intervention” 1.
Biomarkers can be separated into several groups based on a) their characteristics: imaging biomarkers, and molecular biomarkers; b) their applications: diagnostic biomarkers, and drug-related biomarkers 2. They can include specific cells, molecules, genes, gene products, enzymes and hormones.
Molecular biomarkers are biomarkers that were discovered and analyzed by genomics and proteomics. Both genomics and proteomics (as well as transcriptomics {analysis of RNA transcripts}) are combined into Proteogenomics. In addition, other platforms used for the molecular biomarkers identification include: metabolomics (analysis of metabolites), lipidomics (analysis of lipids), glycomics (analysis of polysaccharides) (Figure 1). Despite biomarkers ability to trace environmental contaminants, our major interest lies in using them for the evaluation of the disease status. Every biological system has its own specific biomarkers. Many of us went through routine medical examination, which included the measurement of the level of biomarkers, such as cholesterol or glucose. However, the most important role of biomarkers is the role they play in the diagnosis and prediction or prognosis of diseases, as well as in serving as a target for drugs development.
Figure 1. Biomarkers identification.MS, Mass Spectrometry (including LC-MS, SELDI-TOF, MALDI-TOF, etc.); NMR, Nuclear Magnetic Resonance spectroscopy; MRM, Multiple Reaction Monitoring; SAGE, Serial Analysis of Gene Expression; SDS-PAGE, Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis; 2D-PAGE,2 Dimensional-Polyacrylamide Gel Electrophoresis; WB, Western blot.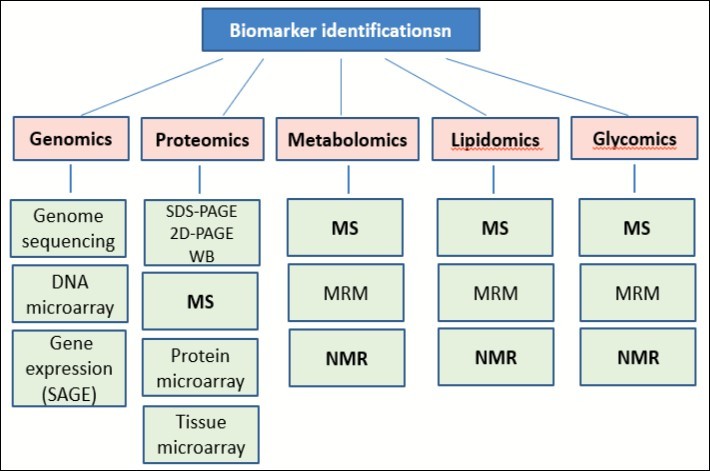
At present, many biomarkers are detected and used in clinical practice but new ones still need to be investigated. Finding new biomarkers is especially important for “complex” diseases with non-clear or multifactorial etiology, such as cancer, neurodegenerative, or gastrointestinal diseases. Let’s take a quick look at the biomarkers for these diseases.
Cancer**
A typical genomic panel for cancer diagnosis is included: BRAF, KIT, NPAS, GNA11, GNAQ for melanomas; ALK, EGFR, KRAS, BRAF for lung cancers; FLT3-ITD/FLT3-TKD, NPM1, c-KIT, PTPN11, CEBPA for acute myeloid leukemia 3. These genes can be used for the diagnosis as well as for targeted drug therapy. For example, EGFR mutations are target for Gefitinib (trade name - Iressa) therapy in non-small cell lung cancer.
** Special issue “Biomarkers for early diagnosis and prognosis of cancer” is planned for publication in JPGR
Neurodegenerative Diseases
As indicated by C. Lausted and colleagues, “most neurodegenerative diseases are characterized by specific abnormal modifications, i.e., misfolded protein aggregates” 4, which could be promising biomarkers: particularly, for Alzheimer’s disease: CSF***-A β42, CSF-A β42/ A β40 (amyloid-beta peptides), CSF-tau, CSF-APP isoforms (amyloid precursor protein), CSF-BACE1 (β-site APP-cleaving enzyme 1); for Parkinson’s disease: CSF a-synuclein, CSF-DJ-1; for Huntington’s disease: CSF-clusterin 4.
*** CSF, cerebrospinal fluid
Gastrointestinal Diseases
Inflammatory Bowel Disease (IBD) is represented a group of chronic intestinal disorders with complex etiology. A number of factors called IBD interactome includes immunome (immune system), microbiome (gut microbiota), exposome (environmental factors), and genome (genetic composition) 5, but practically it manifests as gut inflammation. The best indicator of this process is fecal calprotectin, which is commonly used as molecular biomarkers for IBD diagnosis (in ELISA {Enzyme-Linked Immunosorbent Assay} kits) as addition to the standard endoscopy test. Other evaluated biomarkers (Lactoferrin, ASCA/ANCA {anti-S.cerevisiae antibodies/perinuclear anti-neutrophil cytoplasmic antibodies}, CRP {C-reactive protein}, NGAL {neutrophil gelatinase-associated Lipocalin}) have limited application so far 6.
Optimal biomarkers have to be safe, cost efficient, consistent, modifiable with treatment, easy to obtain and measurable 2. The assay for biomarkers should be rapid, robust, easy to carry out, accurate, inexpensive, and highly specific. Molecular (or nano-) biosensors are new promising tools 7. It is difficult to find the optimal biomarkers for each disease, but this is the primary goal. In many situations a complex of the best biomarkers and techniques may prove to be extremely useful. For example, combination of microRNA and a-synuclein (as well as metabolomics) may bring significant progress in the diagnosis and control of treatment of Parkinson’s disease 8. In any case, early diagnosis of the chronic disorders, such as degenerative diseases and cancer, will help tremendously.
