Abstract
Background
Colonization with methicillin-resistant Staphylococcus aureus (MRSA) is recognized as an association towards development of infections that may cause of morbidity among people living with Human Immunodeficiency Virus (PLWHIV). We report on the prevalence, antibiotic susceptibility pattern and risk factors associated with MRSA carriage among PLWHIV at Nyenga hospital, Buikwe district in central Uganda.
Materials and Methods
We conducted a cross-sectional study among PLWHIV attending Nyenga hospital anti-retroviral therapy (ART) clinic. Nasopharyngeal swab was collected from each participant, cultured to isolate Staphylococcus aureus, and drug susceptibility testing (DST) performed. Sociodemographic data and medical history was recorded.
Results
We enrolled 219 PLWHIV; of these, 58.4% (N=128) were females. The majority of participants (95.0%) were on ART. Ninety-eight (44.75%) of the nasopharyngeal swabs had growth, of which 41 (41.84%) were S. aureus. Of these, 11 (5.02%, 95% confidence interval: 3.67-7.02) were MRSA. Of 41 isolated S. aureus strains, only 8 (19.51%) were susceptible to all antibiotics tested. A total of three (7.32%) were multi-drug resistant (MDR), while one1 (2.43%) was a possible extensively drug resistant (XDR) strain. Deteriorating immunologic state as indicated by a low CD4 count showed a significant association with the MRSA colonization.
Conclusion
These results are reassuring that MRSA colonization is high among PLWHIV. As most of the antibiotics in use were resistant, it raises concerns of intricate clinical management in a low resource set up.
Author Contributions
Academic Editor: HamidReza Naderi, Mashhad University of Medical Sciences
Checked for plagiarism: Yes
Review by: Single-blind
Copyright © 2018 Benedict Ssenyonga, et al.
 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests
The authors have declared that no competing interests exist.
Citation:
Background
Staphylococcus aureus is associated with both nasocomial and community acquired infections 1. It is categorized as either methicillin-resistant S. aureus (MRSA), or methicillin-susceptible, based on the antimicrobials a strain is susceptible to in vitro 2. MRSA poses a public health challenge, in part because of its commensal nature on human skin, and mucous membranes 3, 4. As such, MRSA is readily passed from person-to-person, especially in hospitals), and is often difficult to eradicate 5, 6, 7. At the same time, Antibiotic pressure has led to regular and unwittingly demands for antibiotic use, which has increased the risk of methicillin resistance 8, 9. MRSA strains acquire resistance to beta-lactams and aminoglycosides, the common empirical regimens in sub-Saharan Africa 10, 11. Knowledge of locals antimicrobial susceptibility pattern is essential to select appropriate therapy; however lack of culture and susceptibility testing at most health facilities in resource-limited settings has resulted in widespread empiric therapy. Due to budgetary constraints and lack of programs supporting antimicrobial stewardship, screening for MRSA carriage among PLWHIV in Uganda is not done. This study reports the prevalence, risk factors and DST of asymptomatic MRSA nasal carriage among PLWHIV attending ART clinic at Nyenga hospital in central Uganda.
Methods
Study Design, Area and Population
This cross-sectional study was conducted among PLWHIV attending Nyenga hospital ART clinic between July and November, 2017. Nyenga hospital is a Catholic-founded, private, community not-for-profit facility located in Nyenga town in central Uganda, approximately 15 kilometers west of Jinja. It receives patients from the districts of Buikwe, Jinja, Buvuma and Mukono. The study enrolled consented HIV seropositive clients who were ³18 years of age.
Specimen Collection and Analysis
All study participants had their bilateral anterior nares swabbed with a sterile Dacron dual-swab (Copan Italia Brescia, Italy) which was labeled with a unique identifier. Swabs were transported in a cooler box maintained at 2-80C to the teaching laboratory of Clarke International University (Formerly, International Health Sciences University) for analysis. The dual swab was removed, allowed to equilibrate to room temperature (23-250C) and inoculated directly onto blood agar and MacConkey agar for 24hrs (Fisher, Leicestershire, United Kingdom). After 24 hours, the cultures were checked for growth, and results were recorded as positive or negative. The positive cultures were classified by colony size and morphology, color, and zones of clear beta hemolysis on the media as described 12, 13, 14. A Gram stain was prepared from growth isolates consistent with Staphylococcus species. Biochemical testing was then performed, and Gram-positive cocci that were catalase positive were presumptively identified as Staphylococcus aureus. To confirm bacterial identity, a 0.5 McFarland suspension of the single colony was prepared and then streaked onto CHROMagar-MRSA (BD Diagnostics, Inc.), an MRSA-selective medium. For those samples that grew on CHROMagar-MRSA, additional biochemical identification using coagulase test, DNase test and manitol salt agar was done as described 12, 15, 16. DST was performed by sub culturing a suspension equivalent to 0.5 McFarland standards on to Mueller Hinton Agar (MHA) 17, 18. In vitro drugs tested were penicillin (10µg), gentamycin (10 µg), erythromycin (15 µg), clindamycin (2 µg), vancomycin (30 µg), and cefoxitin (30 µg). Cefoxitin (30 µg) was used as a surrogate marker for MRSA as described in other studies 19, 20, 21. All testing was performed using a standard operating procedure and positive control MRSA strain ATCC 33591 and MSSA strain ATCC 25128.
Data Analysis
Summary statistics were used to describe the cross-sectional cohort, and Chi squared test was used to measure association between variables. Logistic regression analysis was used to determine risk factors associated with MRSA colonization in this population. For all analyses, aP-value of less than 0.05 was considered statistically significant.
Ethical Consideration
Ethical approval was sought from the research and ethics committee of Clarke International University (Formerly called International Health Sciences’ University). Written informed consent was obtained from study participants.
Results
We enrolled 219 PLWHIV, and 58.4% (N=128) were female. The majority of participants were aged 26 to 35 years, (n=86, 39.27%), and 95.0%) were on ART (Table 1).
Table 1. Socio-demographic characteristics of study participants| Variable | Frequency | Percentage |
|---|---|---|
| Age group (Years) | 63 | 28.77 |
| 18-25 | 86 | 39.27 |
| 26-35 | 33 | 15.07 |
| 36-49 | 37 | 16.89 |
| Above 49 | ||
| Education level | ||
| None | 51 | 23.29 |
| Primary | 83 | 37.9 |
| Secondary | 69 | 31.51 |
| Tertiary | 16 | 7.31 |
| CD 4 + cell counts (/ Litre ) | ||
| 0 – 199 | 84 | 38.36 |
| 200 - 499 | 132 | 60.27 |
| Above 500 | 3 | 1.37 |
| Taking ART | ||
| Yes | 208 | 95.0 |
| No | 11 | 5.0 |
| WHO Clinical stage | ||
| I | 41 | 18.72 |
| II | 78 | 35.62 |
| III | 55 | 25.11 |
| IV | 45 | 20.55 |
There were 98 (44.75%) cultures which exhibited growth. Of those, 41 (41.84%) isolates were of Staphylococcus aureus. Of these, 11 participants were found to have MRSA as measured by resistance to Cefoxitin, giving a prevalence of 5.02% (95% confidence interval: 3.67-7.02). Microbiologic results are described in Figure 1.
Figure 1.Microbiologic results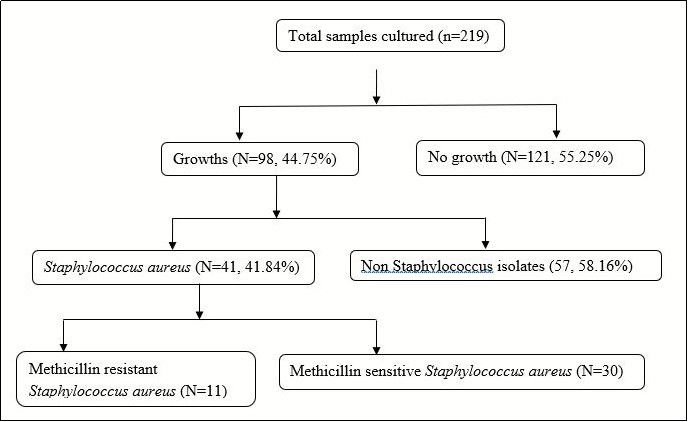
Analysis of the factors associated factors indicated that low CD4 cell counts and deteriorating World Health Organization (WHO) clinical stages were significantly associated with the risk of MRSA colonization. Participants whose CD4 cell counts were less than 500 cells/litre (p<0.05) and WHO clinical stages II (p=0.000), III (p=0.012) and IV (p=0.001) were statistically associated with MRSA colonization, as given in Table 2.
Table 2. Socio-demographic and medical factors associated with MRSA colonization| MRSA colonization | |||
|---|---|---|---|
| Variable | Absent, n= (%) | Present, n= (%) | P-value (95% CI) |
| Age group (Years) | |||
| 18-25 | 62 (29.81) | 1 (9.09) | 1 |
| 26-35 | 82 (39.42) | 4 (36.36) | 0.531 (1.011-1.306) |
| 36-49 | 27 (12.98) | 6 (54.55) | 0.600 (0.576-1.621) |
| Above 49 | 37 (17.79) | 0 (0.0) | 0.150 (0.162-0.197) |
| Education level | |||
| None | 45 (21.63) | 6 (12.9) | 1 |
| Primary | 79 (37.98) | 4 (36.36) | 0.300 (0.248-1.454) |
| Secondary | 79 (37.98) | 4 (36.36) | 0.080 (0.373-1.804) |
| Tertiary | 15 (7.21) | 1(9.09) | 0.210 (0.159-2.062) |
| CD 4 + counts (/ Litre ) | |||
| 0 - 199 | 76(36.54) | 8(72.73) | 0.000*(0.059-0.676) |
| 200 - 499 | 129(62.02) | 3.(27.27) | 0.003* (0.055-0.456) |
| Above 500 | 3(1.44) | 0(0.00) | 1 |
| WHO clinical stage | |||
| I | 40 (19.23) | 1 (9.09) | 1 |
| II | 75 (36.06) | 3(27.27) | 0.000* (0.018-0.171) |
| III | 53 (25.48) | 2 (18.18) | 0.012* (0.023-0.122) |
| IV | 40 (19.23) | 5 (45.45) | 0.001* (0.000-0.164) |
| CI denotes Confidence Interval; 1 is the reference group | |||
DST testing revealed that of 41 isolated S. aureus strains, only 8 (19.51%) were sensitive to all the antibiotics tested, including penicillin. A total of 3 (7.32%) were multi-drug resistant, while 1(2.43%) was a possible XDR in regard of the antibiotics used. This study has revealed that all Staphylococcus aureus was susceptible to vancomycin 41/41 (100%), than clindamycin 24/41 (58.54%), cefoxitin 20/41 (48.78%), erythromycin 14/41 (34.15%) and penicillin 8/41 (19.51%). The susceptibility pattern for clindamycin indicated that 3/17 (17.65%) were inducible resistances, while 14/17 (82.35%) were constitutively resistant. The resistance to erythromycin was 2/46 (4.35%) among all Staphylococcus aureus isolates. The MRSA isolates were more sensitive to vancomycin with 100% sensitivity, while the rest of the antibiotics were poor to gentamycin, clindamycin, erythromycin and penicillin which all gave 100% resistance to MRSA isolates, as presented in Table 3.
Table 3. Antibiotic susceptibility patterns of S. aureus isolates tested to each antibiotic| Drug agent | Strain | Resistant strains | Sensitive strains | Odds ratio | p-value |
|---|---|---|---|---|---|
| Vancomycin (30µg) | MRSA | 0 | 11(26.83) | ||
| MSSA | 0 | 30 (73.17) | |||
| Clindamycin (2µg) | MRSA | 11 (26.83%) | 0 | 1.32 | 0.162 |
| MSSA | 2 (4.88%) | 28 (68.29%) | |||
| Erythromycin (15µg) | MRSA | 11 (26.83%) | 0 | 1.98 | 0.388 |
| MSSA | 16 (39.02%) | 14 (34.15%) | |||
| Gentamycin (10µg) | MRSA | 11 (26.83%) | 0 | 1.69 | 0.511 |
| MSSA | 2 (4.88%) | 28 (68.29) | |||
| Penicillin (10µg) | MRSA | 11 (26.83%) | 0 | 0.27 | 0.102 |
| MSSA | 22 (53.66%) | 8 (19.51%) |
Discussion
To the best of our knowledge, and search, this is the first report of MRSA colonization among PLWHIV attending Nyenga Hospital in Buikwe district. The prevalence of MRSA reported here was 5.02% (95% confidence interval: 3.67-7.02), comparable to the 5.1% reported in Singapore 22, and slightly higher than 2% that was reported in Spain 23 and 2.4% in northern Ethiopia 24. On the other hand, the prevalence is lower than 15.4% reported among an adult cohort from Johns Hopkins University AIDS Service in Baltimore 25, and 16.8% in Northeast Ethiopia 26. The low prevalence of MRSA colonization in this study is ascribed to the occasional visit of PLWHIV to the health facility, since repeated visits or contact with hands of health workers in HIV infected individuals is the major risk factor for colonization 24, 25.
Low CD4 cell counts (less than 200 (p=0.000) and less than 500 cells/litre (p=0.003)) and deteriorating WHO clinical stages were significantly associated with MRSA colonization. This finding affirms to results from previous studies 5, 25. Although mechanisms of resistance were unable to be further characterized, it is notable that HIV-infected clients with advanced immune suppression were more likely to carry MRSA. This is consistent with what was reported from other studies 5, 26, 27.
The assessment of the drug susceptibility pattern of the MRSA isolates indicated high rates of co-resistance of MRSA to commonly prescribed antibiotics such as gentamycin, clindamycin, erythromycin and penicillin which all gave 100% resistance. This is similar to earlier reports 4, 28. The observed high rates of resistance to penicillin, gentamycin, erythromycin, clindamycin, and vancomycin suggest that most of this colonization were caused by resistant strains 28, 29. The high rates of concomitant drug resistance to the commonly available reserve antibiotics for use among the HIV population is of critical attention as S. aureus is a common pathogen among PLWHIV 26. As all participants were ambulatory, those colonized by MRSA could easily transmit the pathogenic bacteria to the community. Our study findings subject to the following limitations, including that nares are not the only primary body site of colonization, so our reported prevalence could under-detect MRSA colonization in this population. In addition, we were unable to genetically characterize MRSA isolates due to resource constraints.
Conclusions:
Our results are reassuring that the overall MRSA colonization is high in our setting. As the majority of the antibiotics in use were resistant, it raises significant concerns of very complicated clinical management. Deteriorating immunological state as measured by low CD4 cell count and advanced clinical stages of infection were significantly associated with MRSA colonization. Premised on these, we have demonstrated the need to encourage routine MRSA screening because of the associated factors.
Acknowledgements
The authors express our utmost gratitude to our participants who voluntarily consented to be enrolled in this study. We are grateful to research assistants for the field, laboratory and data retrieval support.
References
- 1.Shi L. (2017) et al.,Nosocomial and Community-Acquired Spontaneous Bacterial Peritonitis in patients with liver cirrhosis in China: Comparative Microbiology and Therapeutic Implications.Scientific Reports. 7, 46025.
- 2.Enright M C. (2002) The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proceedings of the National Academy of Sciences 99(11), 7687-7692.
- 3.Rasmussen R V.et al.,(2011)Future challenges and treatment of Staphylococcus aureus bacteremia with emphasis on MRSA.Future microbiology. 6(1), 43-56.
- 4.J L Raygada, P D.Levine.(2009)Methicillin-Resistant Staphylococcus aureus: A Growing Risk. in the Hospital and in the Community.American Health & Drug Benefits 2(2), 86-95.
- 5.Hidron A I.et al (2010)Methicillin-resistant Staphylococcus aureus in HIV-infected patients.Infection and drug resistance. 3, 73-86.
- 6.H A Khan, Ahmad A, Mehboob R. (2015) infections and their control strategies.Asian. , Pacific Journal of Tropical Biomedicine 5(7), 509-514.
- 7.Mootsikapun P. (2007) (2007)Bacteremia in adult patients with acquired immunodeficiency syndrome in the northeast of Thailand.International Journal of Infectious Diseases. 11(3), 226-231.
- 8.Loomba P S, Taneja J, B.Mishra (2010)Methicillin and Vancomycin Resistant S. aureus in Hospitalized Patients.Journal of Global Infectious Diseases. 2(3), 275-283.
- 9.Stapleton P D.and PW Taylor (2002)Methicillin resistance in Staphylococcus aureus: mechanisms and modulation.Science progress.85(Pt1):. 57-72.
- 10.MMB Nagelkerke.(2017)Prevalenceof antimicrobial drug resistant bacteria carried by in- and outpatients attending a secondary care hospital in. , Zambia.BMC Research Notes 10, 378.
- 11.PCM Williams, Isaacs D, Berkley J A.(2018)Antimicrobial resistance among children in sub-Saharan Africa.The Lancet. Infectious Diseases. 18(2), 33-44.
- 12.Boerlin P.(2003)Methods for identification of Staphylococcus aureus isolates in cases of bovine mastitis.J. , Clin Microbiol 41(2), 767-71.
- 13.Kaito C, K.. Sekimizu (2007)Colony Spreading in Staphylococcus aureus.Journal of Bacteriology 189(6), 2553-2557.
- 14.Owens W E.and SC Nickerson(1989)Morphologic study of Staphylococcus aureus L-form, reverting, and intermediate colonies in situ.Journal of Clinical Microbiology. 27(6), 1382-1386.
- 15.Kateete D P.(2010)Identification of Staphylococcus aureus: DNase and Mannitol salt agar improve the efficiency of the tube coagulase test.Annals of Clinical Microbiology and Antimicrobials. 9(1), 23.
- 16.Cunha Ribeiro de Souza da.. M.d.L.,Chapter 6 - Methods for the Identification, Characterization, and Tracking the Spread of Staphylococcus aureus A2 -Fetsch, Alexandra, inStaphylococcus aureus2018,AcademicPress 105-125.
- 17.Balouiri M, M. (2016) Sadiki and SK Ibnsouda (2016)Methods for in vitro evaluating antimicrobial activity: A review.Journal of Pharmaceutical Analysis. 6(2), 71-79.
- 18.Qi C, C W Stratton, X.Zheng,Phenotypic testing of bacterial antimicrobial susceptibility. In Advanced Techniques in DiagnosticMicrobiology..2006.SpringerUS.(pp 63-83.
- 19.Fernandes C J.(2005)Cefoxitin resistance as a surrogate marker for the detection of methicillin-resistant Staphylococcus aureus.Journal of Antimicrobial Chemotherapy. 55(4), 506-510.
- 20.Mathews.A et al (2010)Evaluation and comparison of tests to detect methicillin resistant S. , aureus</i>.Indian Journal of Pathology and Microbiology 53(1), 79-82.
- 21.Júnior Sousa.FCd et al (2010)Evaluation of different methods for detecting methicillin resistance in Staphylococcus aureus isolates in a university hospital located in the Northeast of. , Brazil.Brazilian Journal of Microbiology 41, 316-320.
- 22.Kyaw W M.et al (2012)Prevalence of and risk factors for MRSA colonization in HIV-positive outpatients. in Singapore.AIDS Research and Therapy 9, 33-33.
- 23.Imaz.A et al (2015)Prevalence of methicillin-resistant Staphylococcus aureus colonization in HIV-infected patients in Barcelona, Spain: a cross-sectional study.BMC Infectious Diseases.15:. 243.
- 24.Gebremedhn G.et al (2016)Prevalence and risk factors of methicillin-resistant Staphylococcus aureus colonization among HIV patients in Mekelle. , Northern Ethiopia.SpringerPlus 5(1), 877.
- 25.Farley J E.et al (2015)Prevalence and Risk Factors for MRSA in an HIV-positive Cohort.American journal of infection control. 43(4), 329-335.
- 26.Lemma M T.et al (2015)MethicillinResistant Staphylococcus aureus among HIV Infected Pediatric Patients. in Northwest Ethiopia: Carriage Rates and Antibiotic Co-Resistance Profiles.PLoS ONE 10(9), 0137254.
- 27.Stryjewski M E, Corey G R.(2014)Methicillin-Resistant Staphylococcus aureus: An Evolving Pathogen.Clinical Infectious Diseases. 58, 10-19.
