Abstract
GAGA-binding proteins in plants are encoded by the BARLEY B-RECOMBINANT / BASIC PENTACYSTEINE (BBR/BPC) family, which can be spilt into several groups on the basis of sequence divergence. The proteins of the different groups share an evolutionary conserved BASIC PENTACYSTEINE (BPC) domain at their very C-terminus that is important for DNA binding. Hallmark of this domain are five Cysteines at defined positions and spacing, which are considered to form a zinc-finger like structure that is involved in GAGA-motif recognition. Here, we report the formation of stabile homodimers between Arabidopsis thaliana group I member BPC1 or between group II member BPC6 in SDS-PAGE. Serial mutations of the highly conserved five Cysteines in the BPC domain of Arabidopsis thaliana BPC1 were tested for their capacity to bind to GAGA-motifs by DPI-ELISA. Our results do not support the idea of a direct involvement of these residues in making physical contact with the DNA, e.g. by formation of a zinc-finger structure. Instead, the data implies an indispensable function for the five Cysteines in homodimerization and stabilization of the protein structure by disulfide bonds. Accordingly, protein folding and structure prediction suggests the formation of a scaffold for dimerization that is supported by three intermolecular and one intramolecular S-S bond. The high degree of conservation between the BPC domains from the different groups and from different species denotes that this role for the five Cysteines might be evolutionary retained.
Author Contributions
Academic Editor: Laura Scrano, University of Basilicata, Potenza, Italy
Checked for plagiarism: Yes
Review by: Single-blind
Copyright © 2017 Marius L. Theune, et al.
 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests
The authors have declared that no competing interests exist.
Citation:
Introduction
GAGA-motif binding factors (GAF) are indispensable eukaryote transcription factors that act through diverse molecular mechanism during growth and development on homeotic gene expression. Trithorax-like (Trl) and Pipsqueak (Psq) protein families are the representatives of animal GAFs, which possess polyvalent functions in activation and repression of gene expression 1, 2, 3, 4, 5, 6. These proteins affect nucleosome positioning and maintain nucleosome-free chromatin, can cause TATA-proximal pausing of RNA-Polymerases, function as boundary elements or act in silencing of gene expression by interaction with histone-modifying complexes 1, 2, 3, 4, 5, 6, 7, 8. One hypothesis is that these diverse functions of animal GAFs rely on explicit protein-protein interactions with partners that confer process-dependent specificities 3, 9. For example, both Trl and Psq are involved in the sequence-specific recruitment of Polycomb Repressive Complex 1 (PRC1) or PRC2 components to Polycomb Repressive Elements (PREs), which leads to trimethylation of Lys27 in Histone 3 (H3K27me3) 4, 5, 6, 9, 10, 11, 12, 13.
In plants, the three groups of the BARLEY B-RECOMBINANT/BASIC PENTACYSTEINE (BBR/BPC) protein family show GAGA-motif binding properties 14, 15, 16, 17. They are important developmental regulators that are involved in gene expression control mainly of transcription factor genes and influence ethylene, cytokinin, abscisic acid or auxin signaling 18, 19, 20, 21. Although plant and animal GAFs constitute unrelated protein families 14, 16, 22, it was hypothesized that their mode of action at PRE-like motifs might display similarities 15. Indeed, the mechanism of repression of homeotic genes displays striking functional homology: Just like Trl or Psq in animals, the plant BBR/BPC proteins play a role in gene silencing by interaction with repressive complexes. For example, Arabidopsis thaliana group II member BPC6 recruits LIKE-HETEROCHROMATIN PROTEIN 1 (LHP1) to GAGA-motif containing PREs in the promoters of homeotic genes 18. This interaction presumably leads to an association of BPC6 and LHP1 with VERNALIZATION 2 (VRN2), which is a component of the Polycomb Repressive Complex 2 (PRC2) and responsible for H3K27me3 establishment 18, 23. These findings might explain that GAGA-motifs are associated with repressive H3K27me3 modifications close to the transcription start sites 15, 18, 24, 25, 26. Likewise, A. thaliana BPC1, a group I BBR/BPC member, interacts with the SEUSS (SEU) - LEUNIG (LUG) transcriptional cosuppressor complex to control the repression of the homeotic SEEDSTICK (STK) locus 27. Recent data suggest a direct interaction of PRC2 component SWINGER (SWN) with BBR/BPC members of both groups, to repress the expression of the transcription factor gene ABSCISIC ACID INSENSITIVE 4 (ABI4) 20. These repressive mechanisms depend on the specific recruitment of protein complexes to defined chromatin loci in the plant genomes by different groups of BBR/BPC members. Hence, a functional analysis of the DNA-binding mechanism is important to understand the specificity of this recruitment.
The different BBR/BPC groups differ by their structural features at the N-terminus and the central region of the proteins, which presumably harbor dimerization domains or nuclear and nucleolar localization signals 15, 16, 17. Only the BPC DNA-binding domain at the very C-terminus of the proteins is evolutionary conserved and displays a high degree of sequence similarity that extends over the group’s boundaries 15, 16, 22. The hallmark of that BPC domain is the presence of five Cysteine residues, which are highly conserved in position or spacing 15, 16, 17. These Cysteines are believed to form a zinc-finger like structure and to make direct physical contact with the GAGA-tetranucleotide 17. The purpose of this study was to examine the function of these five conserved Cysteines in Arabidopsis thaliana BPC1 and their role in GAGA-motif recognition by DPI-ELISA or protein structure prediction.
Materials and Methods
Protein Expression
The open reading frames of BASIC PENTACYSTEINE 1 (BPC1; AT2G01930) and BPC6 (AT5G42520) were amplified by PCR from a cDNA-library from Arabidopsis thaliana flowers without a stop codon for subsequent cloning into pENTR/D-TOPO (Invitrogen). Site directed mutagenesis of the BPC1 wildtype sequence was essentially performed according to the SPRINP protocol 28. Truncations of the reading frames were generated by PCR. After sequencing, the specific insert was recombined via Gateway (Invitrogen) LR-reaction into the appropriate destination vector pET32b-GW 29, 30. This vector provides translational fusion of a 6xHis-epitope to the N-terminus of the proteins of interest 30. Expression of recombinant proteins was performed in Escherichia coli BL21/RIL cells. Extraction of the total native proteins in the crude soluble fraction was performed according to the DPI-ELISA protocol 29. As a negative control, protein extracts from untransformed E.coli BL21/RIL cells were included in the study.
SDS-Polyacrylamide Gel Electrophoresis and Western Blotting
For protein detection, the crude extracts were first separated by SDS-PAGE in a Mini-PROTEAN Tetra Vertical Electrophoresis Cell (BioRad). Standard cast 10% SDS Mini gels (BioRad) were prepared according to the manufacturer’s description, using Rotiphorese-30 polyacrylamide gel mixtures (Roth). Stacking gels contained 0.125 M Tris (pH 6.8), 0.1% (w/v) SDS, 0.04% (v/v) TEMED, and 0.4% (w/v) APS. Separating gels contained 0.375 M Tris (pH 8.8), 0.1% (w/v) SDS, 0.025% (v/v) TEMED, and 0.125% (w/v) APS. Separating and stacking gels were each allowed to polymerize for 1 h at room temperature. For sample preparation, native protein extracts were mixed with a Laemmli sample buffer (62.5 mM Tris (pH 6.8), 4% SDS, 20% glycerol, 0.01% bromophenol blue, 5% β-mercaptoethanol) and heated for 10 min at 95°C in a bench-top thermo-mixer prior to loading. Electrophoresis was performed at 100 V in Tris-Glycine SDS running buffer (250 mM Tris, 1.92 M Glycine, 1% SDS, pH 8.3) 31. The migration position of the Spectra Multicolor Broad Range Protein Ladder (Fermentas) molecular weight standard was used for mass estimation (kDa). Protein gels were subsequently blotted using a Mini Trans-Blot Cell transfer apparatus (BioRad) and PVDF membranes (Millipore) according to the manufacturer’s descriptions. Membranes were immediately transferred to a Blocking Reagent (Qiagen) or 5% non-fat dry milk (Roth) in TBS-T and incubated with a gentle agitation for about 1 h at room temperature. Immunological detection of 6xHis-epitope tagged proteins was conducted by using mouse anti-His primary antibody (Qiagen)(1/2000), followed by chromogenic detection with alkaline phosphatase (AP)-conjugated rabbit anti-mouse (Qiagen)(1/5000) and NBT/BCIP solution (Roche) according to the manufacturer’s description. Each protein extract was analyzed at least twice independently
DPI-ELISA
DNA binding properties of BPC1 wildtype and mutated proteins were determined using the DPI-ELISA method 29, 32. The amount of total recombinant protein input was normalized for equal loading. For each binding assays, 3 µg total protein in buffered solution and 2 pmol biotinylated oligonucleotides were used per well. The positive (K4) and negative (Kneg) binding dsDNA-probes were used previously 29, 33. DNA binding was studied under oxidizing or reducing conditions by modifications to the protein extraction buffer or the protein dilution buffer 29: To establish an oxidizing environment during protein binding, a final volume of 5% (v/v) H2O2 (Acros Organics) from a fresh bottle was added to the protein dilution buffer prior to the DPI-ELISA. To study the reducing capacity of dithiothreithol (DTT), the usual amount of 5 mM DTT was reduced to 0 mM DTT in only one or in both of the buffers. As BBR/BPC proteins were proposed to constitute possible zinc-finger proteins, DNA binding was analyzed after addition of 100µM ZnCl2 (Sigma) or 100µM EDTA (Sigma) to the protein dilution buffer. Each DPI-ELISA experiment was repeated several times and on different ELISA-plates, with 2-4 technical replicates per plate. Total protein from at least two independent extractions was used. A Tecan Safire plate reader was used for photometric detection. Statistical analysis was performed in Microsoft Excel (MS Office 2013) and VassarStats (http://vassarstats.net/; 2016).
Protein Folding and Structure Prediction
For 3D protein structure prediction the sequence of the BASIC PENTACYSTEINE domain of BPC1 (aa185-aa283) was loaded into I-TASSER 34, 35, 36, 37. The PDB file of the selected model was subsequently used as input for the standalone version of FoldIt 38, 39, 40, 41. The tools ‘repack’ and ‘minimize all’ were applied to the monomeric structure (settings: behavior: start at 0.3 - end at 1.0; wiggle power: medium). After several rounds of relaxation, the derived 3D structure was imported as PDB file into PyMOL (http://www.pymol.org). To generate a dimeric model structure, the monomer was duplicated in PyMol. Each monomer of the resulting dimer model was turned and positioned towards each other. Both molecules were exported as PDB file and again imported into FoldIt. ‘Rubber bands’ were applied to the opposing Cysteine residues. To influence unwanted moving in the process, ‘freeze’ was applied to the entire dimer model, except for the four amino acids surrounding the opposing Cysteine residues. These residues were brought close to each other with the „drag“ tool to enable the formation of disulfide bonds. ‘Rubber bands’ were removed and several rounds of ‘repack’ and ‘minimize all’ (behavior: start at 0.3 - end at 1.0; wiggle power: medium) was applied. The dimer model structure was entirely ‘unfrozen’ and several rounds of „repack“ and „minimize“ were applied on the „unfreeze all’ (wiggle power: medium). The setting behavior was gradually increased (start at 0.3 - end at 1.0) to prevent breaking of the freshly formed bonds. The resulting dimer model structure was exported as PDB file and imported into PyMOL for illustration.
Results
The Arabidopsis thaliana BBR/BPC family consists of three groups with seven members in total 15, 16, 17. The similarities between these proteins are restricted to the BASIC PENTACYSTEINE DNA-binding domain 17. Sequence alignment between A. thaliana group I member BPC1 and BPC6, a group II member, discloses 75 % similarity and 56 % identity over the 99 amino acid long BPC DNA-binding domains (Figure 1A). Two regions of the BPC domains are of highest conservation: On the one hand side, the N-proximal part containing the five conserved Cysteines. On the other hand, a sequence at the very C-terminus, which displays a WA R/K HGTN motif at its center. To investigate the DNA-binding properties by DPI-ELISA, we expressed BPC1 and BPC6 as recombinant His-epitope tagged proteins. By immunological analyses of the crude total protein extracts, we found that both BPC1 and BPC6 formed stabile dimers in SDS-PAGE (Figure 1B). Specific signals exclusively at the molecular weight of the dimers were evident. No signals were detectable at molecular weight of the monomers. We next performed DPI-ELISA experiments with both protein extracts. A strong binding to the positive dsDNA-probe K4 was detected for both recombinant proteins (Figure 1C). No significant interaction with the negative control dsDNA-probe Kneg was observed. Control extracts from BL21/RIL cells and control wells without dsDNA-probe showed only minor background signals. Given the strong homotypic dimerization (Figure 1B), one can propose that exclusively BPC-dimers interacted with GAGA-tetranucleotide motifs in the DPI-ELISA.
Figure 1.Comparison of group I BPC1 and group II BPC6. (A) Protein sequence alignment of the BASIC PENTACYSTEINE DNA-binding domains. Identical residues are highlighted by grey background. Positions that are evolutionary retained in all BBR/BPC family members are indicated by asterisks (*) above the alignment. The highly conserved Cysteines are emphasized by black background. The conserved WA R/K HGTN signature is indicated by red letters. (B) Gel-blot experiments with immunological detection of the recombinant proteins. The expected molecular weights for monomer (*) and dimer (**) proteins are indicated. Arrows point to unspecific bands detected also in control extracts with anti-His antibody. (C) Specific binding of epitope tagged BPC1 and BPC6 to positive (K4) and negative (Kneg) dsDNA-probes in DPI-ELISA experiments. The histogram bars show normalized signal intensities and error bars represent one standard deviation. Grey background shading indicates level of confidence for significant binding (t-test p < 0.05). Representative wells of the microtiter plate are shown below the graph for visual inspection.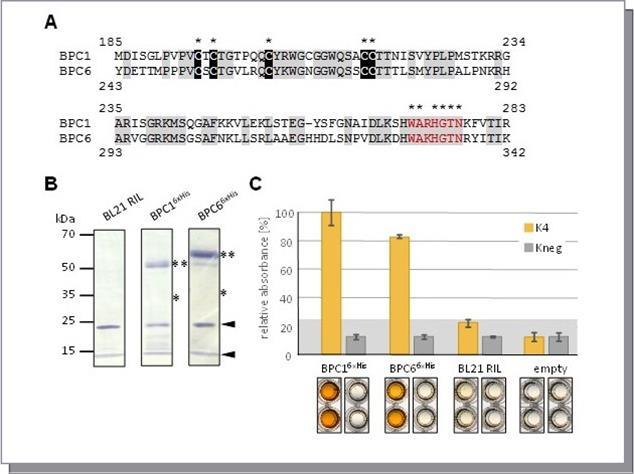
A characteristics of all BBR/BPC proteins is the BASIC PENTACYSTEINE domain that is crucial for DNA-binding 17. The main features of this domain are the highly conserved Cysteines (Figure 2A). To examine the contribution of the five Cysteines in DNA-binding, we conducted site-directed mutagenesis on Arabidopsis thaliana BPC1 to replace selected Cysteines with Glycine. Six different mutant versions of BPC1 were made. In addition, we generated two truncations of BPC1 (Figure 2B): BPC1_DBD comprises the conserved BPC domain (aa185-aa283), while BPC1_short contains only the C-proximal part of the domain starting just after the fifth conserved Cysteine (aa218-aa283). All BPC1 versions were expressed as recombinant His-epitope tagged proteins, which can be detected by immunoblotting (Figure 2C). Interestingly, all signals were approximately double the expected size, which is indicative of putative dimer formation. Also both of the truncations displayed a much higher molecular weight than expected for the monomers.
The DPI-ELISA experiment uncovered that simultaneous mutation of Cys195 and Cys197 (BPC1_mut1) did not affect DNA-binding capacity significantly compared with wildtype BPC1 (Figure 2D). In contrast, the replacement of the single Cys204 with Glycine (BPC1_mut2) reduced binding drastically, which is indicative of an important contribution of this Cysteine to GAGA recognition. Only a minor decrease in binding to GAGA-motifs was found with BPC1_mut3, where Cys216 and Cys217 were mutated. A similar, but insignificant decrease was discovered with a mutation of a Cysteine, which does not belong to the conserved PENTACYSTEINEs (BPC1_mut4). Surprisingly, a huge increase in signal intensity was repeatedly detected with BPC1_mut5 extracts. Here, four of the conserved Cysteines were mutated and only C204 was retained. The result, however, is complementary to the very low binding data with extract BPC1_mut2. Consistently, BPC1_mut6 was derived by mutation of Cys204 from BPC1_mut5 and displayed very low binding affinity to the dsDNA-probes that is similar to BPC1_mut2. Interestingly, BPC1_DBD and BPC1_short did not exhibit any significant binding signals, which suggest an underestimated contribution of regions outside of the conserved BPC domain on GAGA-motif binding. Furthermore, our findings do not support the idea of a direct involvement of all five conserved Cysteines in making physical contact with the DNA.
Figure 2.Binding capacity of BPC1 mutants. (A) Versions of BPC1 with mutations of the Cysteines in the BASIC PENTACYSTEINE DNA-binding domain. (B) Schematic overview of all 6xHis-epitope tagged BPC1 mutants and truncations. The highly conserved Cysteines are highlighted by yellow boxes. The position of the conserved WA R/K HGTN signature is indicated (red). Mutations in Cysteines are shown as crosses. (C) Gel-blot experiments with immunological detection of all recombinant proteins. (D) Specific binding of 6xHis-epitope tagged BPC1 versions to positive (K4) and negative (Kneg) dsDNA-probes in DPI-ELISA experiments. The histogram bars show normalized signal intensities and error bars represent one standard deviation. Grey background shading indicates level of confidence for significant binding (t-test p < 0.05). The bars annotated with the same letter are not significantly different.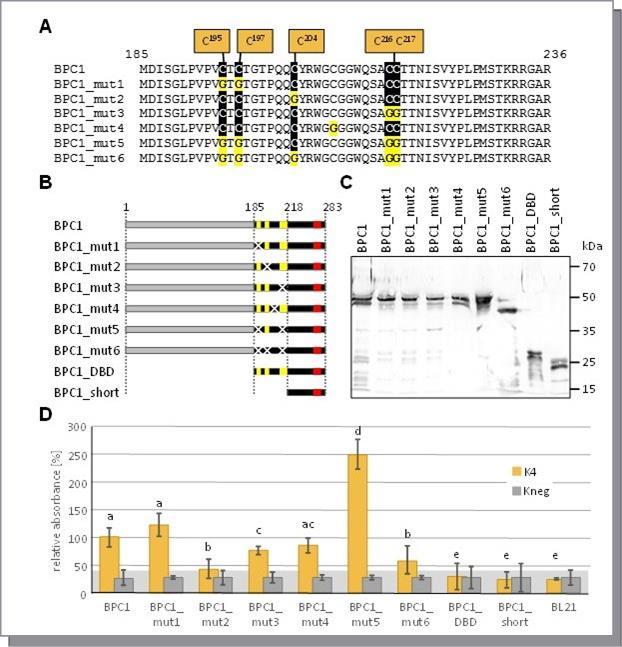
To elucidate the significance of the disulfide bonds in a possible BPC1 dimer for DNA-binding, we conducted a series of DPI-ELISA experiments under reducing or oxidizing buffer conditions. We, therefore, supplemented the protein dilution buffer, which is used for protein binding to the dsDNA-probes in the DPI-ELISA experiments, with the oxidizing agent H2O2 immediately before incubation on the plate. While the protein extracts of the BPC1 variants under investigation displayed consistent DNA-binding capacities under normal buffer conditions, the addition of H2O2 to the protein dilution buffer resulted in a strong reduction of the signal by 40% to 50%
(Figure 3A). Except for BPC1_mut5, the three other BPC1 variants exhibited the same residual, but significant binding activity over the background under oxidizing conditions. This residual activity corresponds to BPC1_mut6 extracts under both conditions, which underlines the significance of the five conserved Cysteines for dimerization and, thus, DNA-binding. As BPC1_mut4 and BPC1 exhibited very similar binding affinities under both conditions, we can exclude an involvement of C209 in DNA-binding or dimer formation. It is noteworthy, however, that the oxidizing conditions led to a general reduction of the signal also with negative binding dsDNA-probes or BL21 control extracts (Figure 3A). We next analyzed the reducing capacity of DTT on DNA-binding by DPI-ELISA (Figure 3B). Similar to the previous experiment, DNA-binding dropped by 40% to 50%, if no reducing agent was included in the protein extraction buffer. In vast contrast, no decrease in binding was found when DTT was lacking in the protein dilution buffer. These results indicate that dimerization must have occurred already prior to the protein extraction inside the E.coli cells. The loss of reducing capacity during protein binding, however, led to a surprising increase in signal in the negative binding dsDNA-probe Kneg (Figure 3B). These data suggest an important role for dimer formation via intermolecular disulfide bonds in the specificity of binding site recognition. To rule out an involvement of the PENTACYSTEINEs in zinc ion complexation, we performed a final DPI-ELISA experiment with an access of Zn2+ ions or in the presence of the chelator EDTA in the protein dilution buffer (Figure 3C). With both supplemented buffers, no difference in binding was observed compared to the control experiment.
Figure 3.Binding capacity of BPC1 under oxidizing or reducing buffer conditions. Specific binding of 6xHis-epitope tagged BPC1 to positive (K4) or negative (Kneg) dsDNA-probes in DPI-ELISA experiments. The histogram bars show normalized signal intensities and error bars represent one standard deviation. Grey background shading indicates level of confidence for significant binding (t-test p < 0.05). The bars annotated with the same letter are not significantly different. (A) Binding capacity under oxidizing conditions. Protein dilution buffer was supplemented with 5% (v/v) H2O2 immediately before the experiment. Asterisks indicate a significant decrease of binding (t-test p < 0.001) by ~ 45%. (B) Analysis of reducing capacity of DTT on DNA-binding. The amount of 5 mM DTT was reduced to 0 mM DTT in the protein extraction buffer (DTTextr.) or the protein dilution buffer (DTTbind.) or in both of the buffers. (C) Effect of 100µM ZnCl2 or 100µM EDTA supplements on DNA-binding.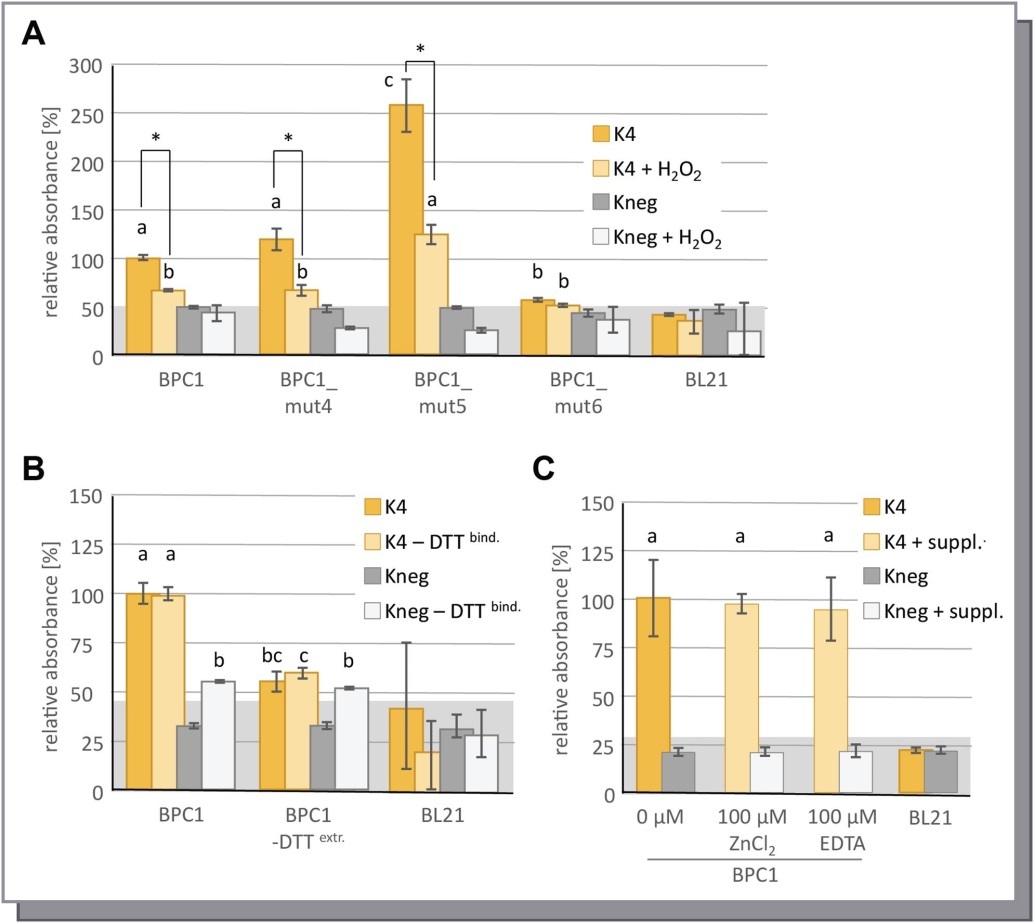
We next used protein structure prediction to derive a 3D model for the BASIC PENTACYSTEINE domain of BPC1. The primary sequence of the domain was loaded into I-TASSER to derive a lead model. We decided for one model (Supplemental Data 1) that was most consistent with simple secondary structure analyses and displayed a consistent beta-beta-alpha-beta signature. This predicted model structure of the BPC domain was loaded into FoldIt for relaxation and subsequently imported into PyMOL for illustration (Supplemental Data 2). The monomeric structure model uncovered that most of the conserved residues of the BPC domain reside at accessible positions at the domain’s surface (Figure 4). Especially, the conserved WA R/K HGTN signature at the C-terminus of the domain is proposed to form a distinctive protrusion. In contrast, the conserved five Cysteines are predicted to be positioned close to each other and to be embedded in rather flat surface area.
Figure 4.Model structure of the BPC1 DNA-binding domain. The monomeric protein model structure was predicted from I-TASSER, relaxed in FoldIt and surface was illustrated in PyMOL. Cysteines at the surface are highlighted in yellow color. The five conserved Cysteine residues are depicted as ribbons. The conserved WA R/K HGTN signature is shown in red color. The intramolecular disulfide bond between Cys204 and Cys217 is indicated.
Such a localization of the Cysteines is not consistent with the idea of the formation of a zinc-finger like structure. In contrast, the 3D model implies that the PENTACYSTEINEs might form a scaffold for interaction via disulfide bonds, which is in agreement with the formation of stabile homotypic dimers. We, therefore, constructed an artificial homodimer on the basis of the monomeric 3D model structure (Figure 5A; Supplemental Data 3). The monomers nicely fit together when the Cysteines of each monomer were opposing each other in a parallel dimer. In fact, three of the Cysteines were able to form intermolecular disulfide bonds in our predicted dimer model. In addition, one additional intramolecular disulfide bond was proposed to form. Closer inspection uncovered that Cys217 – Cys217 and Cys197 – Cys195 pairs are involved in three intermolecular S - S bonds (Figure 5B), which probably have a strong impact on the protein-protein interaction and stabile dimer formation. One intramolecular disulfide bond is predicted between Cys204 and Cys216 that will have stabilizing effects on the entire structure of the BPC domain.
Figure 5.Proposed dimer model structure of two interacting BPC1 DNA-binding domains. (A) Two monomer models were organized as homotypic dimers in PyMOL. Structural modeling and relaxation on the dimer model structure was performed in FoldIt. The surface of each monomer is shown in green or in blue. Cysteines at the surface are highlighted in yellow color. The five conserved Cysteine residues are depicted as ribbons. The conserved WA R/K HGTN signature is shown in red color. (B) Close-up view on the inter- and intramolecular disulfide bonds that are formed in the structural model. Ribbons and labels are displayed in either green or blue to specify the contribution of the monomers. Disulfide bonds are shown in yellow.
Discussion
Here we have shown that the conserved Cysteines in the BPC domain are probably not involved in zinc-finger formation. The analysis of the serial mutations by DPI-ELISA disclosed that some Cysteines appear dispensable for DNA-binding, which suggests no direct involvement in GAGA-motif recognition. Nevertheless, some mutations affected DNA-binding affinity drastically. In line with stabile dimer formation, we propose that the five conserved Cysteines form inter- and intramolecular disulfide bonds in parallel oriented dimers, which are required for specific DNA-binding.
The analysis of all recombinant his-epitope tagged proteins exhibited inconclusive double bands at the approximate sizes of the dimers. One explanation might be degradation products or even rarely occurring posttranslational modifications in E.coli. A closer inspection uncovered that the larger molecular weight band displayed a stronger signal intensity compared with the lower band. A stronger signal for the low molecular weight band, however, was observed for the two constructs BPC1_mut6 and BPC1_short, which lack the five conserved Cysteines. Therefore, we speculate that the double bands somehow represent differentially reduced and oxidized states of some of the disulfide bonds in the BPC domain, which might have occurred during native extraction prior to the addition of reducing agent DTT. In addition, it was noted before, that proteins with extended alpha-helices display a tendency to migrate at higher or lower molecular weight than expected 42, 43, 44. Even considerably minor changes were found to cause drastic alterations in the motility in SDS/PAGE analyses 43. As extended helical structures are predicted for all BBR/BPC family members 15, 16, 17, slightly different protein conformation might already cause altered micelle formation and, hence, will affect the migration in the SDS/PAGE 43. Although the nature of the double bands could not be resolved, the protein extracts provided reproducible and conclusive binding information.
The majority of the protein extracts exhibited a significant and specific binding to GAGA-containing dsDNA-probes. This notion already contradicts the idea that the five Cysteines might directly be involved in DNA-binding via a novel zinc-complexing finger-like structure 17. In addition, our experiments with Zn2+ or EDTA supplemented buffers did not support the idea of a metal chelating role for the five Cysteines. Most noteworthy, however, is the finding that the conserved region of the DNA-binding domain alone (BPC1_DBD) showed no binding, which suggest a regulatory contribution of other parts of the protein that are not contained in the selected BPC domain. Similar results were also found for Arabidopsis thaliana BPC2 in yeast transactivation assays 17, where the DNA-binding domain alone could not induce significant reporter activity. A regulatory function of domains distant from the actual DNA-binding domains is also known for other transcription factors. For example, the large enzymatic ß-amylase domain of BZR-BAM transcription factors is able to communicate with the distal DNA-binding domain and to affect binding affinity 45.
The binding information that converges at Cys204 displays a drastic difference between the mutant versions of BPC1. The single Cys204 versus Gly204 exchange in BPC1_mut2 decreased binding drastically, which is indicative of a possible impact on the entire domain structure. This assumption was consolidated by our model structure, as intramolecular disulfide bonds were predicted for Cys204 and Cys216. Similarly, the replacement of all five Cysteines (BPC1_mut6) caused a very weak binding to DNA and possibly reflect binding affinities of BPC1 monomers. Consistently, the oxidizing agent H2O2 did not affect BPC1_mut6 binding, because this mutant version already mimics an oxidized state. To our surprise, the effects of the simultaneous mutation of Cys216 and Cys217 (BPC1_mut3), which also deletes one of the partners for intramolecular disulfide bonding, were not comparable to BPC1_mut2 and rather mild. Moreover, the replacement of all conserved Cysteines except for Cys204 displayed strongest signal intensities (BPC1_mut5). Keeping in mind that DPI-ELISA provide semi-quantitative data of high comparability, 29, 32, 33, we must conclude that a mutation in Cys204 affects binding affinity maybe through an altered flexibility of the protein. As we always used the same amount of dsDNA-probe with an identical number of accessible GAGA-tetranucleotides in all experiments, the mutations in BPC1_mut5 must have decreased binding specificities. Hence, binding to other sequences besides the GAGA-motif would be permissive and more proteins could possibly bind per dsDNA-probe, which translates into increased signal intensities. This assumption was supported by our experiment with the reducing agent DTT, where an increased binding to the negative binding dsDNA-probe was found under non-reducing conditions. Interestingly, all other observations concerning the binding experiments can readily be explained by the 3D model structure and by an alternated retaining of the intermolecular disulfide bonds.
A dimerization in parallel conformation places the structural protrusions of the conserved WA R/K HGTN signature in the focus for further DNA-binding studies. This region is basic and exhibits a partial positive charge. Hence, this signature motif might be involved in making direct contact with the acidic backbone of the DNA. Interestingly, this short WA R/K HGTN stretch of BBR/BPC proteins is reminiscent of the WRKYGQK signature of WRKY transcription factors 30, 46, 47, 48. Both sequences are probably contained in beta-sheets that possibly protrude into the major groove of the DNA to make contact to nucleotides at both sides of the structures 16, 46, 47. In addition, the distribution and position of large and small or polar and aliphatic amino acid residues displays surprising similarities between these seemingly unrelated protein families.
The probable interaction as parallel oriented dimers suggest that two neighboring GAGA tetranucleotide motifs are bound, which is consistent with previous reports 16. It remains elusive, however, whether or not BBR/BPC monomers are capable of binding to DNA at all. This study data and previous reports 17 demonstrate that the BPC DNA-binding domain alone is not able to bind to GAGA-motifs. This might not be the case in planta, where probably several proteins contribute to the specificity of binding. Nevertheless, contacting two neighboring GAGA-motifs discloses important functional implications on target loci identification by increasing binding specificity by a 256 fold. Possible binding to longer GAGA-consensi is consistent with the findings that the repressive H3K27me3 mark of PRC2 is enriched at extended GA/TC-dinucleotides in PREs in plants 18, 24, 26. Also a genome wide analysis of GA/TC-dinucleotides uncovered a significant bias towards longer GAGA-motifs in promoter and in intron sequences 15, 25.
The formation of homotypic dimers in Arabidopsis thaliana BBR/BPC proteins was reported before. Group II member BPC6 has been shown to dimerize with the aid of an Alanine-zipper interaction domain, which is localized in the N-terminal part of the protein 16. Opposing positive and negative charged residues form salt bridges that support a strong interaction of parallel dimers in vivo and in silico. Transient bimolecular fluorescence complementation (BiFC) experiments with a BPC6 version that lacks the C-terminus in Nicotiana benthamiana demonstrated that the BPC DNA-binding domain and its conserved Cysteines were not required for homotypic dimerization 16. Spectro-microscopic analysis with FRET-FLIM consolidated a parallel dimer conformation in vivo16, which is consistent with our data presented in this manuscript on the parallel orientation of the BPC-domain dimers, which are essential for disulfide bond formation. Experiments with group I member BPC1 discovered bands that were shifted and supershifted in EMSA, which is an indication for dimer formation in vitro17, 20, 49. Unfortunately, the use of longer DNA-probes with extended GA/TC-repeats might give inconclusive results, as a single GAGA-tetranucleotide is sufficient for BBR/BPC-binding in vitro29, 33: The simultaneous binding of several BBR/BPC proteins independently to the same DNA-probe or the binding of dimers to neighboring sites might provide identical outcomes 17, 20, 33, 49. All these data suggest, however, the formation of higher order complexes of BBR/BPC proteins at GAGA-motifs.
The high degree of conservation of the five Cysteines in all BBR/BPC family members proposes a conserved mechanism. In the light of our recent findings, we propose that these residues might possibly constitute a general interaction surface between BBR/BPC proteins, which implies a possible dimer formation also between members of different groups. Indeed, there is preliminary data that heterotypic dimerization between different group members might occur. A weak but significant heterodimer formation between Arabidopsis BPC1 and BPC6 was found in yeast two-hybrid experiments 16. This interaction, however, could not be consolidated in BiFC experiments in planta16. In contrast, spectro-microscopic analyses with ectopically overexpressed BPC1 and BPC6 in heterologous Nicotiana benthamiana cells suggest a very close association of both proteins in the nucleus 50. The proportion of the two proteins, however, that actually underwent a possible heterotypic dimerization compared with those forming homodimers in planta was not resolved the study. Hence, these reports on heterotypic dimerization between members of different groups might constitute artifacts due to ectopic overexpression. To clarify the dynamics between the different groups of BBR/BPC proteins in the cell and their mechanistic function needs further experiments on these issues in the future.
Conclusion
Our serial analysis of mutants in the conserved BASIC PENTACYSTEINE DNA-binding domain contradicts the idea of a zinc-finger-like DNA-binding mechanistic. Instead, we propose that the conserved Cysteines form a scaffold for homotypic dimerization of BBR/BPC proteins under native conditions. Inter- and intramolecular disulfide bonds stabilize a parallel conformation of the monomers. Such a conformation will consequentially lead to the recognition and binding of neighboring GAGA-motifs, which will have tremendous impact on target loci selection in vivo.
Supplemental Data
Supplemental data 1: PDB-file of the I-TASSER model
Supplemental data 2: PDB-file of the 3D model structure of the monomer
Supplemental data 3: PDB-file of the 3D model structure of the dimer
Acknowledgements
We like to acknowledge Angelika Anna, Luise Brand, Alexander Böser, Jan Hirsch, Pascal Karitter, Nathalie Sebening and Christine Zehren for continuous support and technical assistance. The experimental work was supported by Biomers, Gemany, and through basic funding of the Universität des Saarlandes, the Universität Tübingen and by financial support of the ZHMB.
Copy Right
The authors agree to publication under CREATIVE COMMONS LICENSE.
References
- 1.N L Adkins, T A Hagerman, Georgel P. (2006) GAGA protein: a multi-faceted transcription factor. , Biochem Cell Biol. v 84, 559-567.
- 2.Lehmann M. (2004) Anything else but GAGA: a nonhistone protein complex reshapes chromatin structure. , Trends Genet. v 20, 15-22.
- 3.Lomaev D, Mikhailova A, Erokhin M, A V Shaposhnikov. (2017) The GAGA factor regulatory network: Identification of GAGA factor associated proteins. PLoS One. 12, 0173602.
- 4.Mishra K, V S Chopra, Srinivasan A, R K Mishra. (2003) Trl-GAGA directly interacts with lola like and both are part of the repressive complex of Polycomb group of genes. , Mech Dev. v 120, 681-689.
- 5.N M Mulholland, I F King, R E Kingston. (2003) Regulation of Polycomb group complexes by the sequence-specific DNA binding proteins ZesteandGAGA.Genes Dev. 17, 2741-2746.
- 6.Salvaing J, Lopez A, Boivin A, J S Deutsch. (2003) The Drosophila Corto protein interacts with Polycomb-group proteins and the GAGA factor. , Nucleic Acids Res. v 31, 2873-2882.
- 7.S Y Tsai, Y L Chang, K B Swamy, R L Chiang. (2016) GAGA factor, a positive regulator of global gene expression, modulates transcriptional pausing and organization of upstream nucleosomes. , Epigenetics Chromatin 9, 32.
- 8.N J Fuda, M J Guertin, Sharma S, C G Danko. (2015) GAGA factor maintains nucleosome-free regions and has a role in RNA polymerase II recruitment to promoters. PLoS Genet. 11, 1005108.
- 9.Schuettengruber B, Cavalli G. (2009) Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. , Development. v 136, 3531-3542.
- 10.Faucheux M, J Y Roignant, Netter S, Charollais J. (2003) batman Interacts with polycomb and trithorax group genes and encodes a BTB/POZ protein that is included in a complex containing GAGA factor. , Mol Cell Biol. v 23, 1181-1195.
- 11.H B Li, Ohno K, Gui H, Pirrotta V. (2013) Insulators target active genes to transcription factories and polycomb-repressed genes to polycomb bodies. , PLoS Genet 9, 1003436.
- 12.Schuettengruber B, Cavalli G. (2013) Polycomb domain formation depends on short and long distance regulatory cues. PLoS One. 8, 56531.
- 13.Wang L, Jahren N, E L Miller, C S Ketel. (2010) Comparative analysis of chromatin binding by Sex Comb on Midleg (SCM) and other polycomb group repressors at a Drosophila Hox gene. , Mol Cell Biol. v 30, 2584-2593.
- 14.Sangwan I, M R O’brian. (2002) Identification of a soybean protein that interacts with GAGA element dinucleotide repeat DNA. , Plant Physiol. v 129, 94.
- 15.Santi L, Wang Y, M R Stile, Berendzen K. (2003) The GA octodinucleotide repeat binding factor BBR participates in the transcriptional regulation of the homeobox gene Bkn3. , Plant J. v 34, 813-826.
- 16.Wanke D, M L Hohenstatt, Dynowski M, Bloss U. (2011) Alanine zipper-like coiled-coil domains are necessary for homotypic dimerization of plant GAGA-factors in the nucleus and nucleolus. PLoS One. 6, 16070.
- 17.Meister R J, Williams L A, Monfared M M, Gallagher T L. (2004) Definition and interactions of a positive regulatory element of the Arabidopsis INNER NO OUTER promoter. , Plant J. v 37, 426-438.
- 18.Hecker A, L H Brand, Peter S, Simoncello N. (2015) . The Arabidopsis GAGA-Binding Factor BASIC PENTACYSTEINE6 Recruits the POLYCOMB-REPRESSIVE COMPLEX1 Component LIKE HETEROCHROMATIN PROTEIN1 to GAGA DNA Motifs. Plant Physiol. v. 168 1013-1024.
- 19.Monfared M M, Simon M K, Meister R J, Roig-Villanova I. (2011) Overlapping and antagonistic activities of BASIC PENTACYSTEINE genes affect a range of developmental processes in Arabidopsis. , Plant J. v 66, 1020-1031.
- 20.Mu Y, Zou M, Sun X, He B. (2017) . BASIC PENTACYSTEINE Proteins Repress ABSCISIC ACID INSENSITIVE4 Expression via Direct Recruitment of the Polycomb-Repressive Complex 2 in Arabidopsis Root Development. Plant Cell Physiol. v. 58 607-621.
- 21.Simonini S, Kater M M. (2014) Class I BASIC PENTACYSTEINE factors regulate HOMEOBOX genes involved in meristem size maintenance. , J Exp Bot. v 65, 1455-1465.
- 22.Lang D, Weiche B, Timmerhaus G, Richardt S. (2010) Genome-wide phylogenetic comparative analysis of plant transcriptional regulation: a timeline of loss, gain, expansion, and correlation with complexity. , Genome Biol Evol 2, 488-503.
- 23.Lafos M, Kroll P, M L Hohenstatt, F L Thorpe. (2011) Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. , PLoS Genet 7, 1002040.
- 24.Berke L, Snel B. (2014) The histone modification H3K27me3 is retained after gene duplication and correlates with conserved noncoding sequences in Arabidopsis. , Genome Biol Evol 6, 572-579.
- 25.K W Berendzen, Stuber K, Harter K, Wanke D. (2006) Cis-motifs upstream of the transcription and translation initiation sites are effectively revealed by their positional disequilibrium in eukaryote genomes using frequency distribution curves. , BMC Bioinformatics 7, 522.
- 26.Deng W, D M Buzas, Ying H, Robertson M. (2013) Arabidopsis Polycomb Repressive Complex 2 binding sites contain putative GAGA factor binding motifs within coding regions of genes. , BMC Genomics. v 14, 593.
- 27.Simonini S, Roig-Villanova I, Gregis V, Colombo B. (2012) Basic pentacysteine proteins mediate MADS domain complex binding to the DNA for tissue-specific expression of target genes in Arabidopsis. Plant Cell. 24, 4163-4172.
- 28.Edelheit O, Hanukoglu A, Hanukoglu I. (2009) Simple and efficient site-directed mutagenesis using two single-primer reactions in parallel to generate mutants for protein structure-function studies. , BMC Biotechnol 9, 61.
- 29.L H Brand, Kirchler T, Hummel S, Chaban C. (2010) DPI-ELISA: a fast and versatile method to specify the binding of plant transcription factors to DNA in vitro. Plant Methods. 6, 25.
- 30.Ciolkowski I, Wanke D, R P Birkenbihl, I E Somssich. (2008) Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. , Plant Mol Biol. v 68, 81-92.
- 31.Sambrook J, D W Russell. (2001) Molecular cloning : a laboratory manual, 3rd ed.Cold Spring Harbor,N.Y.:ColdSpringHarborLaboratoryPress.
- 32.Brand L H, Satbhai S B, Kolukisaoglu H U, Wanke D. (2013) Limits And Prospects Of Methods For The Analysis Of DNA-Protein Interaction, in The Analysis of Regulatory DNA: Current Developments, Knowledge and Applications Uncovering Gene Regulation. , Berendzen K W, Wanke DKilian J, Editors. Bentham Science Publishers 124-148.
- 33.S M Fischer, Böser A, J P Hirsch, Wanke D. (2016) Quantitative Analysis of Protein-DNA Interaction by qDPI-ELISA. , Methods Mol Biol 1482, 49-66.
- 34.Yang J, Zhang Y. (2015) I-TASSER server: new development for protein structure and function predictions. , Nucleic Acids Res. v 43, 174-181.
- 35.Yang J, Yan R, Roy A, Xu D. (2015) The I-TASSER Suite: protein structure and function prediction. , Nat Methods. v 12, 7-8.
- 36.Roy A, Kucukural A, Zhang Y. (2010) I-TASSER: a unified platform for automated protein structure and function prediction. , Nat Protoc 5, 725-738.
- 38.C B Eiben, J B Siegel, J B Bale, Cooper S. (2012) Increased Diels-Alderase activity through backbone remodeling guided by Foldit players. , Nat Biotechnol. v 30, 190-192.
- 39.Cooper S, Khatib F, Treuille A, Barbero J. (2010) Predicting protein structures with a multiplayer online game. , Nature. v 466, 756-760.
- 40.Khatib F, Cooper S, M D Tyka, Xu K. (2011) Algorithm discovery by protein folding game players. , Proc Natl Acad Sci U S A. v 108, 18949-18953.
- 41.Khatib F, Dimaio F, Foldit Contenders G, G Foldit Void Crushers. (2011) Crystal structure of a monomeric retroviral protease solved by protein folding game players. , Nat Struct Mol Biol. v 18, 1175-1177.
- 42.Rath A, Cunningham F, C M Deber. (2013) Acrylamide concentration determines the direction and magnitude of helical membrane protein gel shifts. Proc Natl Acad Sci U S A. v 110-15668.
- 43.Rath A, Glibowicka M, V G Nadeau, Chen G. (2009) Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. , Proc Natl Acad Sci U S A. v 106, 1760-1765.
- 44.Walkenhorst W F, Merzlyakov M, Hristova K, Wimley W C. (2009) Polar residues in transmembrane helices can decrease electrophoretic mobility in polyacrylamide gels without causing helix dimerization. , Biochim Biophys Acta 1788, 1321-1331.
- 45.Soyk S, Simkova K, Zurcher E, Luginbuhl L. (2014) The Enzyme-Like Domain of Arabidopsis Nuclear beta-Amylases Is Critical for DNA Sequence Recognition and Transcriptional Activation. Plant Cell. , v 26, 1746-1763.
- 46.Yamasaki K, Kigawa T, Watanabe S, Inoue M. (2012) Structural basis for sequence-specific DNA recognition by an Arabidopsis WRKY transcription factor. , J Biol Chem. v 287, 7683-7691.
- 47.L H Brand, N M Fischer, Harter K, Kohlbacher O. (2013) Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. , Nucleic Acids Res. v 41, 9764-9778.
- 48.Rinerson C I, Rabara R C, Tripathi P, Shen Q J. (2015) The evolution of WRKY transcription factors. , BMC Plant Biol. v 15, 66.
Cited by (6)
- 1.Zhang Shuaiwei, Wang Jinmiao, Feng Yunqiang, Xue Yanxu, Wang Yudan, et al, 2023, Genome-wide identification of flowering Chinese cabbage BPC family genes and BcBPC9 functional analysis in Cd stress tolerance, Plant Stress, 10(), 100220, 10.1016/j.stress.2023.100220
- 2.Shanks Carly M., Hecker Andreas, Cheng Chia‐Yi, Brand Luise, Collani Silvio, et al, 2018, Role of BASIC PENTACYSTEINE transcription factors in a subset of cytokinin signaling responses, The Plant Journal, 95(3), 458, 10.1111/tpj.13962
- 3.Theune Marius L., Bloss Ulrich, Brand Luise H., Ladwig Friederike, Wanke Dierk, 2019, Phylogenetic Analyses and GAGA-Motif Binding Studies of BBR/BPC Proteins Lend to Clues in GAGA-Motif Recognition and a Regulatory Role in Brassinosteroid Signaling, Frontiers in Plant Science, 10(), 10.3389/fpls.2019.00466
- 4.Sahu Anubhav, Singh Ritu, Verma Praveen Kumar, 2023, Plant BBR/BPC transcription factors: unlocking multilayered regulation in development, stress and immunity, Planta, 258(2), 10.1007/s00425-023-04188-y
- 5.Li Shuzhen, Miao Li, Huang Bin, Gao Lihong, He Chaoxing, et al, 2019, Genome-Wide Identification and Characterization of Cucumber BPC Transcription Factors and Their Responses to Abiotic Stresses and Exogenous Phytohormones, International Journal of Molecular Sciences, 20(20), 5048, 10.3390/ijms20205048
- 6.Ahad Arzoo, Aslam Roohi, Gul Alvina, Amir Rabia, Munir Faiza, et al, 2021, Genome-wide analysis of bZIP, BBR, and BZR transcription factors in Triticum aestivum, PLOS ONE, 16(11), e0259404, 10.1371/journal.pone.0259404
